Following the work of Yao et al. (see poster below for reference) in their work on the original CERENKOV model, this project sought to replicate their results while also testing some previously unmentioned methods/models.
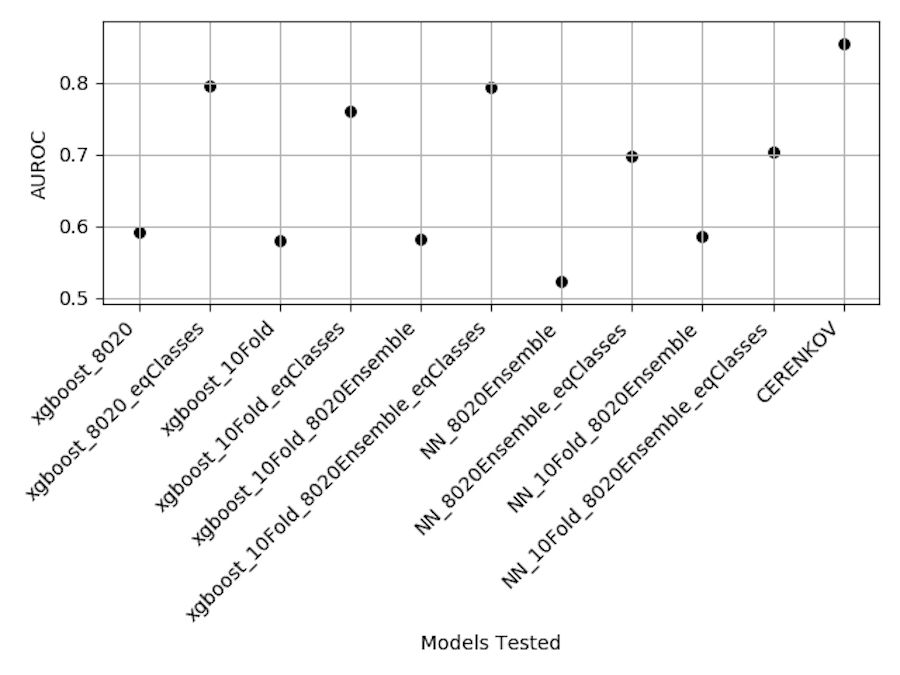
We trained XGBoost and neural network classifiers to predict rSNP in non-coding genome regions. Our methods and results are reviewed in the poster below. Overall, we found that the data sampling method (from an unbalanced dataset) had a greater affect on AUROC than the actual model used (XGBoost vs. NN, with and without k-Fold cross-validation).
The original CERENKOV project used a novel method of locus-based cross-fold validation which we attempted to replicate. Though we didn't quite reach the same amount of performance, we hope that our results might spur more interest in this project, especially using data resampling methods.
Below are different model results in terms of AUROC from our experiments.
The poster can be view here.
For anyone interested in our code/process the source is also available in our GitHub repo.